- Teacher: Marvin ALBERT
- Teacher: Vincent GUILLEMOT
- Teacher: Frédéric LEMOINE
- Teacher: Nicolas MAILLET
- Teacher: Emmanuelle PERMAL
- Teacher: Emeline PERTHAME
Hub Moodle
Available courses
Introduction to R programming
- Teacher: Anthony BERTRAND
- Teacher: Andela DAVIDOVIC
- Teacher: Blaise LI
- Teacher: Sébastien MELLA
Session in French – Confidence intervals and Hypothesis testing: a little bit of theory and practice with R
6/11/2023 – Full day
8/11/2023 – Afternoon
9/11/2023 – Afternoon
- Teacher: Juliette MEYER
- Teacher: Thomas OBADIA
- Teacher: Anthony BERTRAND
- Teacher: Elise JACQUEMET
- Teacher: Hugo VARET
- Teacher: Evangelia ELEFTHERIOU
- Teacher: Vincent LAVILLE
Introduction to R programming
- Teacher: Maëlle DAUNESSE
- Teacher: Andela DAVIDOVIC
- Teacher: Blaise LI
- Teacher: Sébastien MELLA
Session in French – Confidence intervals and Hypothesis testing: a little bit of theory and practice with R
10/04/2025 – Full day
11/04/2025 – Full day
- Teacher: Juliette MEYER
- Teacher: Thomas OBADIA
- Teacher: Elise JACQUEMET
- Teacher: Hugo VARET
- Teacher: Maëlle DAUNESSE
- Teacher: Vincent GUILLEMOT
- Teacher: Vincent LAVILLE
Objectives: knowing how to
-
Navigate the filesystem, identify files by their path, create, move, rename, delete files and directories
-
Extract information from text files
Objectives:
-
Understand biological information available online
-
Understand sequence comparisons using local alignment
-
Find information about unknown sequences, using online tools
This course is intended to provide the students with the prerequisites needed to be able to follow the courses of the Bioinformatics track dedicated to the analysis of NGS data (B4, B5, B6, B7, B8, B9, B10, B11).
The course has three main parts:
1 - Main UNIX commands
2 - Main R commands / getting acquainted with R scripts and Rstudio
3 - Main Galaxy commands
Objectives:
-
Understand the key concepts underlying the quantification and comparison of OMIC datasets
-
Perform the initial steps of most HTS data analysis
Objectives:
-
Design a successful transcriptomics experiment
-
How to get differential expressed genes from raw data
- Teacher: Juliette MEYER
Objectives:
-
Learn the key steps to obtain genetic reliable variants from raw sequencing reads
-
First applications : use filtered files of variants to perform basic analyses
Objectives:
-
Getting exposed to important applications in the analysis of genetic data, genomewide
-
Analysing genomic data with R and other popular software
The courses will introduce several analysis techniques in microbial ecology. Participants will receive an overview of state-the-art tools from bioinformatic processing to the statistical analysis (raw reads processing, metagenomes assembly, contigs annotations, metagenomes binning, samples comparison and OTU processing).
Objectives:
-
Get acquainted with the key concepts of scRNAseq raw data analysis, quantification, functional characterization and visualization.
- Teacher: Sébastien MELLA
Objectives
-
Understand the key concepts of functional analysis
-
Perform functional analysis on RNA-Seq data on a simple design using R tools
Objectives:
-
understand more complex but useful options and commands
-
learn regular expressions and few commands using them,
-
everything needed to create your firsts own scripts.
- Teacher: Julien GUGLIELMINI
- Teacher: Julia Kende
- Teacher: Nicolas MAILLET
Date: Tuesday, January the 8th, 14:00am to 17:30pm.
Where: room 5, building 6, under the canteen.
Teacher: Stéphane Rigaud
Assistant teacher: Marie Anselmet and Gaëlle Letort
This course provides a comprehensive introduction to QuPath, an open-source software platform designed for bioimage analysis of large 2D images, from digital-pathology to large slide scan. Participants will learn the basic of the software, how to navigate and uses QuPath for analyzing images, object annotations, and segmentation.
The course covers essential topics such as project creation, annotation, tissue and nuclei segmentation, and quantification, enabling users to perform quantitative analysis of tissue samples. Using both QuPath integrated tools and State of the Art Deep-Learning extensions.
Course materials:
- A laptop along with a mouse and a power connector
- Download and install QuPath: https://qupath.github.io/
- Download the following datasets:
Additional material: QuPath Documentation
- Teacher: Marie ANSELMET
- Teacher: Gaëlle LETORT
- Teacher: Stephane RIGAUD

This course is an introductory course on utilizing Python for the analysis of biological images. While Fiji is great for quick and user-friendly image processing workflows, Python offers unparalleled flexibility and access to advanced tools like deep learning-based segmentation, batch processing, and custom analysis pipelines tailored to your research needs. The course is meant for students who have no or little experience with Python and basic knowledge in bio-image analysis (for example from following IA1).
- Teacher: Marvin ALBERT
- Teacher: Marie ANSELMET
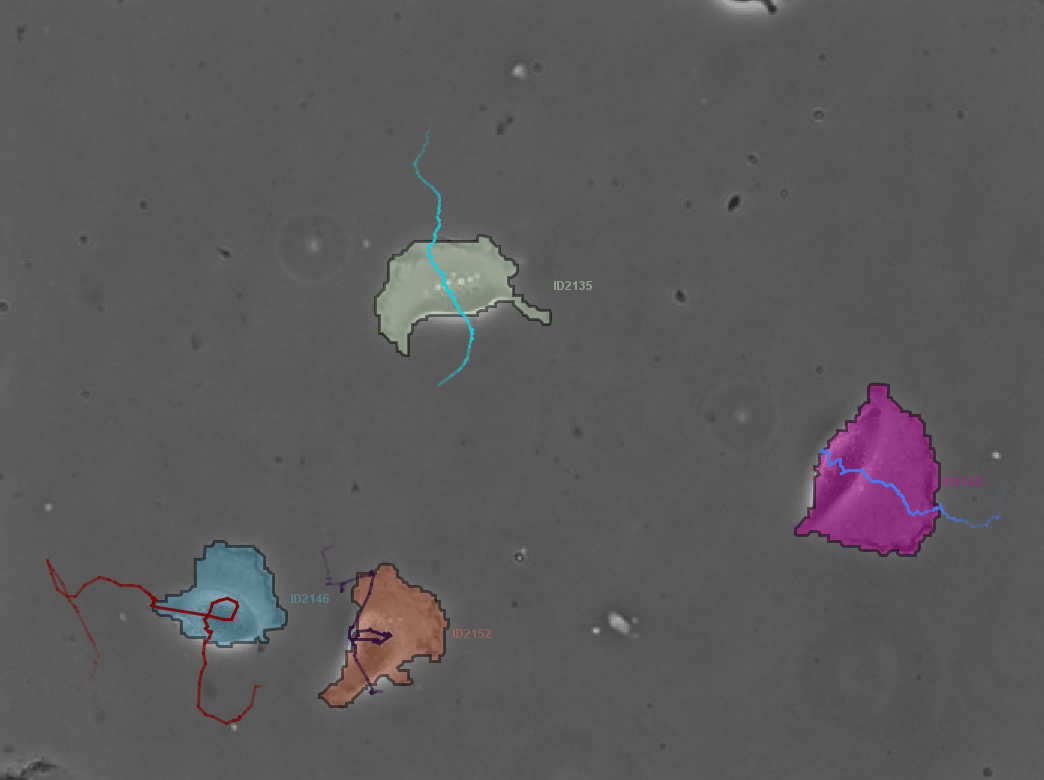
Using TrackMate for analysis of time-lapse microscopy movies.
This course is aimed at beginners in bioimage analysis, that wants to learn how to analyse time-lapse movies. Such movies can follow the movement of cells, in 2D or 3D over time, or the motility of organelles and large objects. They offer unique insight into the dynamics of biology objects, on the evolution of their morphology and of molecular processes, imaged via transmission light or fluorescence microscopy.
This half a day course includes a brief theoretical introduction about tracking algorithms, and is followed by a hands-on tutorial. For the tutorial we will use TrackMate (https://imagej.net/plugins/trackmate/), which is a somewhat popular Fiji plugin for tracking, developed in the Image Analysis Hub.
What do you need to attend:
- Your laptop, Mac or PC
- A mouse (mandatory)
- Fiji installed (https://fiji.sc)
- The following tutorial materials:
https://dl.pasteur.fr/fop/Te8uJcsC/TrackMateTutorialMaterial-2025.zip
For more information contact Jean-Yves.
- Teacher: Jean-Yves TINEVEZ
- Teacher: Minh Son PHAN
- Teacher: Laura XENARD
- Teacher: Minh Son PHAN
- Teacher: Jean-Yves TINEVEZ
- Teacher: Minh Son PHAN
- Teacher: Son Phan
- Teacher: Jean-Yves TINEVEZ
Tutorial on how to use QuPath for large fixed images analysis and in particular histopathological images.
- Teacher: Gaëlle LETORT
- Teacher: Jean-Yves TINEVEZ
- Teacher: Gaëlle LETORT
- Teacher: Jean-Yves TINEVEZ
Cours d'Introduction à la programmation en R
- Teacher: Vincent GUILLEMOT
- Teacher: Yoann Madec
Cours d'Introduction à la programmation en R
- Teacher: Vincent GUILLEMOT
- Teacher: Yoann Madec
- Teacher: Hervé BOURHY
- Teacher: Malika JELLAOUI
- Teacher: Perrine PARIZE
- Teacher: Hervé BOURHY
- Teacher: Malika JELLAOUI
- Teacher: Perrine PARIZE
Description
- Teacher: Claudia CHICA
- Teacher: Bernd JAGLA
- Teacher: Yann LOE MIE
- Teacher: Sébastien MELLA
- Teacher: Olivier MIRABEAU